Selection of minimum CORESET Size for a Given Target Block Probability
In this notebook, we simulate a design problem in determining the minimum CORESET size needed for meeting a target blocking probability.
Specifically, given the number of UEs and the coverage condition, BS can properly determine the CORESET size to ensure the blocking probability does not exceed a specified threshold.
For evaluation, we consider the
medium coveragecondidtion where most of the UEs require medium ALs (i.e., AL 4), with AL distribution [0.05, 0.2, 0.5, 0.2, 0.05]The CORESET size must increase when more UEs are scheduled and a smaller blocking probability target.
While a larger CORESET is beneficial for UE scheduling, it may not be desired from spectral and energy efficiency perspective, as more bandwith and more number of CCEs that a UE has to blindly look for. Therfore the network should properly select the CORESET size based on the requirements. So there is a trade off between CORESET size and target blocking probability.
Python Libraries
[1]:
import os
os.environ["CUDA_VISIBLE_DEVICES"] = "-1"
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'
# %matplotlib widget
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import matplotlib as mpl
import numpy as np
5G-Toolkit Libraries
[2]:
import sys
sys.path.append("../../")
from toolkit5G.Scheduler import PDCCHScheduler
Simulation Parameters
The following parameters are used for this simulation: - coresetID denotes the coreset ID. - slotNumber denotes the slot-number carrying the PDCCH. - searchSpaceType denotes the search space type. UE specific search space (USS) or Common search space (CSS). - nci denotes the variable corresponding to carrier aggregation. Current simulation does not assume carrier aggregation.
[3]:
mu = np.random.randint(4) # numerlogy for sub-carrier spacing
numSlotsPerFrame = 2**mu * 10 # number of slots per radio frame
coresetID = 1 # coreset ID
slotNumber = 0
searchSpaceType = "USS" # search space type. UE specific search space
nci = 0 # variable corresponding to carrier aggregation
numIterations = 1000
PDCCH Scheduling Parameters
Following parameters are crucial for PDCCH scheduling performance: - coresetSize denotes coreset size or number of CCEs available for scheduling UEs. - strategy denotes the scheduling strategy. - numCandidates denotes number of PDCCH candidates per each Aggregation Level (AL). - aggLevelProbDistribution denotes the probability distribution with which an AL is choosen for a UE by the scheduler.
[4]:
aggLevelProbDistribution = np.array([0.05, 0.2, 0.5, 0.2, 0.05])
minCCEs = 20
maxCCEs = 135
coresetSize = np.arange(minCCEs,maxCCEs+1,5)
numCandidates = np.array([135,67,33,16,8], dtype=int)
strategy = "Conservative"
pdcchSchedulerObj = PDCCHScheduler(mu, slotNumber, coresetID, nci)
Compute minimum coreset size for numUEs = 5.
[5]:
################
# For 5 UEs
################
numUEs = 5
rnti = np.random.choice( np.arange(1,65519+1), size = (numUEs,), replace=False)
probOfBlocking = np.zeros((coresetSize.size,))
for n in range(coresetSize.size):
print("Simulating (n,numCCEs) : "+str(n)+", "+str(coresetSize[n]))
Nccep = coresetSize[n]
prob = 0
for i in range(numIterations):
ueALdistribution = np.random.multinomial(numUEs, aggLevelProbDistribution)
count = pdcchSchedulerObj(Nccep,searchSpaceType,ueALdistribution,numCandidates,rnti,strategy)[0]
numBlockedUEs = np.sum(count)
prob = prob + (numBlockedUEs/numUEs)
probOfBlocking[n] = prob/numIterations
targetBlockingProbability = 0.05
minCoresetSize5ForNumUEs5 = coresetSize[np.where(probOfBlocking<=targetBlockingProbability)[0][0]]
targetBlockingProbability = 0.10
minCoresetSize10ForNumUEs5 = coresetSize[np.where(probOfBlocking<=targetBlockingProbability)[0][0]]
targetBlockingProbability = 0.15
minCoresetSize15ForNumUEs5 = coresetSize[np.where(probOfBlocking<=targetBlockingProbability)[0][0]]
targetBlockingProbability = 0.20
minCoresetSize20ForNumUEs5 = coresetSize[np.where(probOfBlocking<=targetBlockingProbability)[0][0]]
Simulating (n,numCCEs) : 0, 20
Simulating (n,numCCEs) : 1, 25
Simulating (n,numCCEs) : 2, 30
Simulating (n,numCCEs) : 3, 35
Simulating (n,numCCEs) : 4, 40
Simulating (n,numCCEs) : 5, 45
Simulating (n,numCCEs) : 6, 50
Simulating (n,numCCEs) : 7, 55
Simulating (n,numCCEs) : 8, 60
Simulating (n,numCCEs) : 9, 65
Simulating (n,numCCEs) : 10, 70
Simulating (n,numCCEs) : 11, 75
Simulating (n,numCCEs) : 12, 80
Simulating (n,numCCEs) : 13, 85
Simulating (n,numCCEs) : 14, 90
Simulating (n,numCCEs) : 15, 95
Simulating (n,numCCEs) : 16, 100
Simulating (n,numCCEs) : 17, 105
Simulating (n,numCCEs) : 18, 110
Simulating (n,numCCEs) : 19, 115
Simulating (n,numCCEs) : 20, 120
Simulating (n,numCCEs) : 21, 125
Simulating (n,numCCEs) : 22, 130
Simulating (n,numCCEs) : 23, 135
Compute minimum coreset size for numUEs = 10.
[6]:
################
# For 10 UEs
################
numUEs = 10
rnti = np.random.choice( np.arange(1,65519+1), size = (numUEs,), replace=False)
probOfBlocking = np.zeros((coresetSize.size,))
for n in range(coresetSize.shape[0]):
print("Simulating (n,numCCEs) : "+str(n)+", "+str(coresetSize[n]))
Nccep = coresetSize[n]
prob = 0
for i in range(numIterations):
ueALdistribution = np.random.multinomial(numUEs, aggLevelProbDistribution)
count = pdcchSchedulerObj(Nccep,searchSpaceType,ueALdistribution,numCandidates,rnti,strategy)[0]
numBlockedUEs = np.sum(count)
prob = prob + (numBlockedUEs/numUEs)
probOfBlocking[n] = prob/numIterations
targetBlockingProbability = 0.05
minCoresetSize5ForNumUEs10 = coresetSize[np.where(probOfBlocking<=targetBlockingProbability)[0][0]]
targetBlockingProbability = 0.10
minCoresetSize10ForNumUEs10 = coresetSize[np.where(probOfBlocking<=targetBlockingProbability)[0][0]]
targetBlockingProbability = 0.15
minCoresetSize15ForNumUEs10 = coresetSize[np.where(probOfBlocking<=targetBlockingProbability)[0][0]]
targetBlockingProbability = 0.20
minCoresetSize20ForNumUEs10 = coresetSize[np.where(probOfBlocking<=targetBlockingProbability)[0][0]]
Simulating (n,numCCEs) : 0, 20
Simulating (n,numCCEs) : 1, 25
Simulating (n,numCCEs) : 2, 30
Simulating (n,numCCEs) : 3, 35
Simulating (n,numCCEs) : 4, 40
Simulating (n,numCCEs) : 5, 45
Simulating (n,numCCEs) : 6, 50
Simulating (n,numCCEs) : 7, 55
Simulating (n,numCCEs) : 8, 60
Simulating (n,numCCEs) : 9, 65
Simulating (n,numCCEs) : 10, 70
Simulating (n,numCCEs) : 11, 75
Simulating (n,numCCEs) : 12, 80
Simulating (n,numCCEs) : 13, 85
Simulating (n,numCCEs) : 14, 90
Simulating (n,numCCEs) : 15, 95
Simulating (n,numCCEs) : 16, 100
Simulating (n,numCCEs) : 17, 105
Simulating (n,numCCEs) : 18, 110
Simulating (n,numCCEs) : 19, 115
Simulating (n,numCCEs) : 20, 120
Simulating (n,numCCEs) : 21, 125
Simulating (n,numCCEs) : 22, 130
Simulating (n,numCCEs) : 23, 135
Compute minimum coreset size for numUEs = 15.
[7]:
################
# For 15 UEs
################
numUEs = 15
rnti = np.random.choice( np.arange(1,65519+1), size = (numUEs,), replace=False)
probOfBlocking = np.zeros((coresetSize.size,))
for n in range(coresetSize.shape[0]):
print("Simulating (n,numCCEs) : "+str(n)+", "+str(coresetSize[n]))
Nccep = coresetSize[n]
prob = 0
for i in range(numIterations):
ueALdistribution = np.random.multinomial(numUEs, aggLevelProbDistribution)
count = pdcchSchedulerObj(Nccep,searchSpaceType,ueALdistribution,numCandidates,rnti,strategy)[0]
numBlockedUEs = np.sum(count)
prob = prob + numBlockedUEs/numUEs
probOfBlocking[n] = prob/numIterations
targetBlockingProbability = 0.05
minCoresetSize5ForNumUEs15 = coresetSize[np.where(probOfBlocking<=targetBlockingProbability)[0][0]]
targetBlockingProbability = 0.10
minCoresetSize10ForNumUEs15 = coresetSize[np.where(probOfBlocking<=targetBlockingProbability)[0][0]]
targetBlockingProbability = 0.15
minCoresetSize15ForNumUEs15 = coresetSize[np.where(probOfBlocking<=targetBlockingProbability)[0][0]]
targetBlockingProbability = 0.20
minCoresetSize20ForNumUEs15 = coresetSize[np.where(probOfBlocking<=targetBlockingProbability)[0][0]]
Simulating (n,numCCEs) : 0, 20
Simulating (n,numCCEs) : 1, 25
Simulating (n,numCCEs) : 2, 30
Simulating (n,numCCEs) : 3, 35
Simulating (n,numCCEs) : 4, 40
Simulating (n,numCCEs) : 5, 45
Simulating (n,numCCEs) : 6, 50
Simulating (n,numCCEs) : 7, 55
Simulating (n,numCCEs) : 8, 60
Simulating (n,numCCEs) : 9, 65
Simulating (n,numCCEs) : 10, 70
Simulating (n,numCCEs) : 11, 75
Simulating (n,numCCEs) : 12, 80
Simulating (n,numCCEs) : 13, 85
Simulating (n,numCCEs) : 14, 90
Simulating (n,numCCEs) : 15, 95
Simulating (n,numCCEs) : 16, 100
Simulating (n,numCCEs) : 17, 105
Simulating (n,numCCEs) : 18, 110
Simulating (n,numCCEs) : 19, 115
Simulating (n,numCCEs) : 20, 120
Simulating (n,numCCEs) : 21, 125
Simulating (n,numCCEs) : 22, 130
Simulating (n,numCCEs) : 23, 135
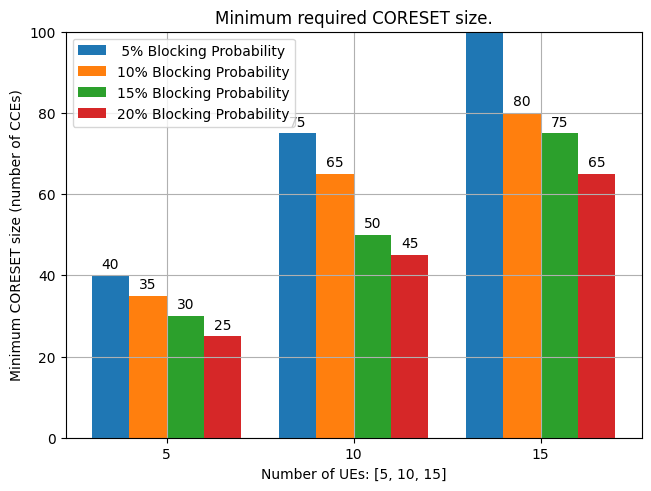
Display Minimum CORESET size required to meet the Target Blocking Probability for different number of UEs.
Its the recreation of
Fig. 11. Minimum required CORESET size for different number of UEs and blocking probability requirementsfrom the reference paper referenced below [1].Figure shows that the minimum required CORESET size for 5, 10, 15 UEs and different blocking probability targets of 5%. 10%, 15% and 20%.
Cleary, we see that the CORESET size increases when more UEs are scheduled and a smaller blocking probability target. For instance, comparing two cases:
case-1: 5 UEs and 20% blocking probability target
case-2: 10 UEs and 15% blocking probability target
It shows that CORESET size for case-2 needs 3 times larger than that of case-1 (i.e. from 25 CCEs to 75 CCEs).
[8]:
species = ("5", "10", "15")
penguin_means = {' 5% Blocking Probability' : (minCoresetSize5ForNumUEs5, minCoresetSize5ForNumUEs10, minCoresetSize5ForNumUEs15),
'10% Blocking Probability': (minCoresetSize10ForNumUEs5, minCoresetSize10ForNumUEs10, minCoresetSize10ForNumUEs15),
'15% Blocking Probability': (minCoresetSize15ForNumUEs5, minCoresetSize15ForNumUEs10, minCoresetSize15ForNumUEs15),
'20% Blocking Probability': (minCoresetSize20ForNumUEs5, minCoresetSize20ForNumUEs10, minCoresetSize20ForNumUEs15)}
x = np.array([3,8,13]) # the label locations
width = 1 # the width of the bars
multiplier = 1
fig, ax = plt.subplots(layout='constrained')
for attribute, measurement in penguin_means.items():
offset = width * multiplier
rects = ax.bar(x + offset, measurement, width, label=attribute)
ax.bar_label(rects, padding=3)
multiplier += 1
# Add some text for labels, title and custom x-axis tick labels, etc.
ax.set_ylabel('Minimum CORESET size (number of CCEs)')
ax.set_xlabel('Number of UEs: [5, 10, 15]')
ax.set_title('Minimum required CORESET size.')
ax.set_xticks(x+2.5, species)
ax.legend(loc='upper left', ncols=1)
ax.set_ylim(0, 100)
ax.grid()
plt.show()
References
[1] Blocking Probability Analysis for 5G New Radio (NR) Physical Downlink Control Channel. Mohammad Mozaffari, Y.-P. Eric Wang, and Kittipong Kittichokechai
[ ]: