Analyze the Impact of UE Capability on Blocking Probability
In this notebook, we simulate the impact of UE capability in terms of Blind Decoding (BD)/ CCE limits on the
blocking probabilityWhen BD/CCE limits is reduced, UE can monitor a fewer number of CCEs per slot. This can also limit the scheduling flexibilty and increases blocking probability.
For evaluation, we consider the following cases:
Reference Case: Assuming that the UE is configured to monitor [6,6,4,2,1] PDCCH candiates for ALs [1,2,4,8,16].
Reduced BD Case A: Assuming that the UE is configured to monitor [3,3,2,1,1] PDCCH candiates for ALs [1,2,4,8,16],. In this case BD limit is reduced by 50% compared to reference case.
Reduced BD Case B: Assuming that the UE is configured to monitor [1,1,1,1,1] PDCCH candiates for ALs [1,2,4,8,16]. In this case BD limit is reduced by around 75% compared to reference case.
Python Libraries
[1]:
import os
os.environ["CUDA_VISIBLE_DEVICES"] = "-1"
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'
# %matplotlib widget
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import matplotlib as mpl
import numpy as np
5G-Toolkit Libraries
[2]:
import sys
sys.path.append("../../")
from toolkit5G.Scheduler import PDCCHScheduler
Simulation Parameters
The following parameters are used for this simulation: - coresetID denotes the coreset ID. - slotNumber denotes the slot-number carrying the PDCCH. - searchSpaceType denotes the search space type. UE specific search space (USS) or Common search space (CSS). - nci denotes the variable corresponding to carrier aggregation. Current simulation does not assume carrier aggregation.
[3]:
mu = np.random.randint(4) # numerlogy for sub-carrier spacing
numSlotsPerFrame = 2**mu * 10 # number of slots per radio frame
coresetID = 1 # coreset ID
slotNumber = 0
searchSpaceType = "USS" # search space type. UE specific search space
nci = 0 # variable corresponding to carrier aggregation
numIterations = 1000
numUEs = 20 # number of UEs in simulation
PDCCH Scheduling Parameters
Following parameters are crucial for PDCCH scheduling performance: - coresetSize denotes coreset size or number of CCEs available for scheduling UEs. - strategy denotes the scheduling strategy. - numCandidates denotes number of PDCCH candidates per each Aggregation Level.
[4]:
minCCEs = 20
maxCCEs = 80
coresetSize = np.arange(minCCEs,maxCCEs+1,10)
strategy = "Conservative"
aggLevelProbDistribution = np.array([0.4, 0.3, 0.2, 0.05, 0.05])
pdcchSchedulerObj = PDCCHScheduler(mu, slotNumber, coresetID, nci)
Simulating the Reference Case
[5]:
#################
# Reference Case
#################
probOfBlockingRefCase = np.zeros(coresetSize.shape)
numCandidates = np.array([6,6,4,2,1], dtype=int)
print("##################################################################################################")
for n in range(coresetSize.size):
Nccep = coresetSize[n]
print("Current Coreset Size:", Nccep)
prob = 0
for i in range(numIterations):
ueALdistribution = np.random.multinomial(numUEs, aggLevelProbDistribution)
rnti = np.random.choice( np.arange(1,65519+1), size = (numUEs,), replace=False)
count = pdcchSchedulerObj(Nccep,searchSpaceType,ueALdistribution,numCandidates,rnti,strategy)[0]
numBlockedUEs = np.sum(count)
prob = prob + numBlockedUEs/numUEs
probOfBlockingRefCase[n] = prob/numIterations
print("##################################################################################################")
##################################################################################################
Current Coreset Size: 20
Current Coreset Size: 30
Current Coreset Size: 40
Current Coreset Size: 50
Current Coreset Size: 60
Current Coreset Size: 70
Current Coreset Size: 80
##################################################################################################
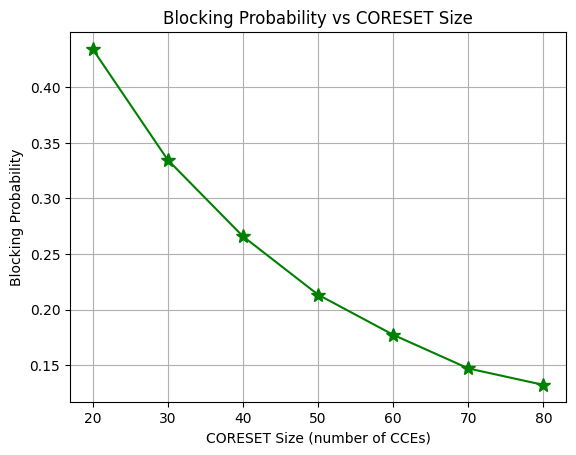
Plot Blocking Probability for Different CORESET Sizes for Different UEs
Its the recreation of
Fig. 5. Blocking probability versus CORESET size (number of CCEs).from the reference paper referenced below [1].Intuitively as the number of CCEs increases, BS scheduler gets more flexibility to allocate non-ovelapping set of CCEs to each UE and hence the blocking probability decreases.
From the figure, we see that the blocking probability can be reduced from 0.19 to 0.04 by increasing the number of CCEs in CORESET from 30 to 60.
Note that the impact of further increasing the CORESET size is minimal as almost all the UEs can be successfully scheduled.
[6]:
fig, ax = plt.subplots()
ax.plot(coresetSize, probOfBlockingRefCase, marker = "*",linestyle = "solid", ms = 10, c = 'g')
ax.set_xlabel('CORESET Size (number of CCEs)')
ax.set_ylabel('Blocking Probability')
ax.set_title('Blocking Probability vs CORESET Size', fontsize=12)
ax.set_xticks(coresetSize)
# ax.set_xlim([coresetSize[0], coresetSize[-1]])
ax.grid()
plt.show()
Simulating Reduced Blind Decoding Case-A
[7]:
#####################
# Reduced BD Case A
####################
probOfBlockingBDCase_A = np.zeros(coresetSize.shape)
numCandidates = np.array([3,3,2,1,1], dtype=int)
print("##################################################################################################")
for n in range(coresetSize.size):
Nccep = coresetSize[n]
print("Current Coreset Size:", Nccep)
prob = 0
for i in range(numIterations):
ueALdistribution = np.random.multinomial(numUEs, aggLevelProbDistribution)
rnti = np.random.choice( np.arange(1,65519+1), size = (numUEs,), replace=False)
count = pdcchSchedulerObj(Nccep,searchSpaceType,ueALdistribution,numCandidates,rnti,strategy)[0]
numBlockedUEs = np.sum(count)
prob = prob + numBlockedUEs/numUEs
probOfBlockingBDCase_A[n] = prob/numIterations
print("##################################################################################################")
##################################################################################################
Current Coreset Size: 20
Current Coreset Size: 30
Current Coreset Size: 40
Current Coreset Size: 50
Current Coreset Size: 60
Current Coreset Size: 70
Current Coreset Size: 80
##################################################################################################
Simulating Reduced Blind Decoding Case-B
[8]:
#####################
# Reduced BD Case B
####################
probOfBlockingBDCase_B = np.zeros(coresetSize.shape)
prevALIndices = np.array([], dtype = int)
numCandidates = np.array([1,1,1,1,1], dtype=int)
print("##################################################################################################")
for n in range(coresetSize.size):
Nccep = coresetSize[n]
print("Current Coreset Size:", Nccep)
prob = 0
for i in range(numIterations):
ueALdistribution = np.random.multinomial(numUEs, aggLevelProbDistribution)
rnti = np.random.choice( np.arange(1,65519+1), size = (numUEs,), replace=False)
count = pdcchSchedulerObj(Nccep,searchSpaceType,ueALdistribution,numCandidates,rnti,strategy)[0]
numBlockedUEs = np.sum(count)
prob = prob + numBlockedUEs/numUEs
probOfBlockingBDCase_B[n] = prob/numIterations
print("##################################################################################################")
##################################################################################################
Current Coreset Size: 20
Current Coreset Size: 30
Current Coreset Size: 40
Current Coreset Size: 50
Current Coreset Size: 60
Current Coreset Size: 70
Current Coreset Size: 80
##################################################################################################
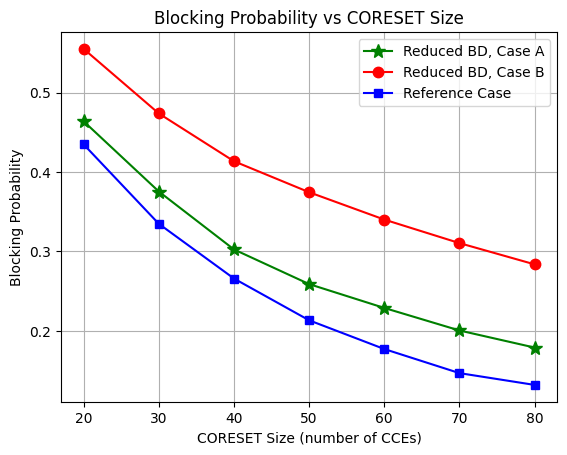
Plot Blocking Probability for Different CORESET Sizes for Different UEs
Its the recreation of
Fig. 9: Blocking probability for different blind decoding (BD) capabilitiesfrom the reference paper referenced below [1].From the figure, we see that the blocking probabilty increase by a factor of 2.4 and 5.6, when reducing the BD limit by 50% and 75% compared to the reference case.
[9]:
fig, ax = plt.subplots()
ax.plot(coresetSize, probOfBlockingBDCase_A, marker = "*",linestyle = "solid", ms = 10, c = 'g',label = "Reduced BD, Case A")
ax.plot(coresetSize, probOfBlockingBDCase_B, marker = "o",linestyle = "solid", ms = 7.5, c = 'r',label = "Reduced BD, Case B")
ax.plot(coresetSize, probOfBlockingRefCase, marker = "s",linestyle = "solid", ms = 6, c = 'b',label = "Reference Case")
ax.legend()
ax.set_xlabel('CORESET Size (number of CCEs)')
ax.set_ylabel('Blocking Probability')
ax.set_title('Blocking Probability vs CORESET Size', fontsize=12)
ax.set_xticks(coresetSize)
# ax.set_xlim([coresetSize[0], coresetSize[-1]])
ax.grid()
plt.show()
References
[1] Blocking Probability Analysis for 5G New Radio (NR) Physical Downlink Control Channel. Mohammad Mozaffari, Y.-P. Eric Wang, and Kittipong Kittichokechai
[ ]: